CoNtRol
This page documents the CoNtRol web interface. It is currently very much a work in progress.
Introduction
CoNtRol (CRN tool) is a web application that allows users to run various tests to determine the possible dynamics of chemical reaction networks. It is intended to be an open framework allowing researchers to build on each other's work by adding new tests as new developments occur. The reference implementation is available at http://reaction-networks.net/control/.
User Documentation
There are a number of ways to use CoNtRol. The most basic way is to type a set of reactions into the web page and view the results of the analysis online. This mode also offers a number of helper functions, such as a Java Web Start application to draw the DSR graph and a function to generate LaTeX describing the reactions. Users can also save the chemical reaction network structure in a text file for future analysis. It is also possible to upload a set of chemical reactions in a variety of file formats.
More advanced users can edit options to disable some of the tests, or to restrict tests to mass action kinetics (for tests that support this restriction). For users with multiple reaction networks to analyse, there is also a feature to upload a batch of networks for analysis, with the results being sent to the user via email.
The individual tools that make up CoNtRol can also be built and run individually on users’ local machines if preferred. The web interface is purely intended to help simplify the use of multiple tools.
Pre-built versions of CoNtRol are available for download at http://reaction-networks.net/control/download/.
TO DO: include documentation about running individual components manually. Link to individual component downloads once they are available.
Basic Usage
The simplest way to use CoNtRol is simply to type a set of reactions into the main input form. Pressing the “Analyse CRN” button will then begin analysis of the CRN and load the results once the calculations are complete. By default, all tests are run, and debugging output is not included. At the bottom of the results are buttons to generate the DSR graph of the CRN, and to email the calculated results for later use.
To remove all reactions and reset any stored results, click the “--” button.
Advanced Usage
CoNtRol offers several advanced options:
- Download/upload of a file describing a CRN (to simplify the re-analysis of a CRN, or to allow input of a CRN from a catalogue of CRNs)
- Upload of a zip archive containing multiple CRN descriptor files
- Enable/disable certain tests
- Turn on/off extra debugging output
- Automatic generation of LaTex markup describing the CRN chemical equations and stoichiometry matrix, and TikZ markup describing the DSR graph
By default, the Analysis menu only shows the “Analyse CRN” and “Options” buttons. The “Options” button allows users to enable/disable tests and debugging (detailed test output) mode. Clicking “More” at the bottom of the Analysis menu offers the ability to download a CRN descriptor file, to generate LaTeX for the reaction equations and stoichiometry matrix, and to launch the DSR graph application.
The File Input menu is hidden by default. Clicking “Show” allows the user to upload either a single file, for analysis within the web page, or a batch file for delayed analysis. In both modes, currently supported CRN file formats are listed. Batch files must consist of a zip archive containing one or more text files representing CRNs, all in the same format.
DSR graph application
CoNtRol includes a Java Web Start application to draw the DSR graph. It can be launched from the Analysis menu, or from the button at the bottom of the results page. The reaction and reactant nodes can be moved around with the mouse. Clicking the “Export to LaTeX” button will open a dialogue to save a file containing TikZ markup describing the DSR graph.
Note: when using Google Chrome/Chromium with default settings, the DSR graph will not launch automatically in Java Web Start. See https://code.google.com/p/chromium/issues/detail?id=10877#c206 for a workaround for this issue.
Running the DSR graph application requires that Java be installed on the user’s computer. Unfortunately this means that the DSR graph feature cannot be used on mobile devices. All other CoNtRol features should work on mobile devices, including Android and iOS smartphones and tablets.
Developer Documentation
The entire CoNtRol application is open source, to promote its re-use and development by as wide a number of researchers as possible. Developers can browse the entire source tree at http://reaction-networks.net/control/download/source. Currently the web application is only supported on Linux servers, although we believe that it will also run on Mac OS X with some custom PHP modules. There are no plans for Windows support at the moment but we believe that it should run under Cygwin (feedback/testing results appreciated). Windows users can still use CoNtRol via a browser, it is only not supported for Windows servers. Additionally, some/all of the individual components should run on Windows.
CoNtRol currently consists of five components, written in different languages. The user interface and server “glue” that launches the tests and fetches results is written in PHP, with the display component written in standards compliant XHTML 1.1/CSS/jQuery/JavaScript. The DSR graph display is a Java Web Start application. The “General analysis“ (analysereacs) test is written in C. The “DSR” (dsrtest) test is written in GNU Octave. The “Jacobian” (calc-jacobian) test is written in Maxima, with some PHP and POSIX shell script wrapper code around it.
New tests can be implemented as plugins in (theoretically) any language, including scripts for proprietary tools such as MATLAB, Mathematica, etc, although they may require some shell scripting glue to fit into the CoNtRol architecture.
Plugin Architecture
New plugins can be added by providing the following:
- An executable file that can be invoked via the command line using the form
plugin [--mass-action-only] filename- The
--mass-action-onlyflag is optional, and is only required for executables that support both mass action and general kinetics. - The executable can either be a compiled binary, or a shell script, which may in turn simply be a wrapper calling other binaries or scripts.
- The
- An array describing the plugin, to be added to
includes/standard-tests.php. Examples are provided.
File Formats
CoNtRol currently supports four file formats, although there is scope to add more in the future if there is a requirement for them. All currently supported file formats are plain text. Samples of each are shown below for the reaction network
Net Stoichiometry
-1 0 0 1 1 -1 1 -1 1
This format is very simple but also very limited, as it does not show whether each reaction is reversible or irreversible, nor does it show if a reactant appears on both sides of a reaction. Currently CoNtRol assumes that all reactions are irreversible and that no reactant appears on both sides of a reaction when this file format is used. To represent a reversible reaction, it is necessary to include both the forwards and backwards reactions.
Net Stoichiometry + V Matrix
S MATRIX -1 0 1 1 1 -1 V MATRIX 1 0 0 2 0 1
This format is slightly less limited as it can distinguish reversible and irreversible reactions. The zeros in the first column of the V matrix show that the reaction is irreversible. By convention, a 2 in the V matrix represents ±, i.e. the reaction rate can depend positively or negatively on the reactant concentration at different points in phase space.
Source Stoichiometry + Target Stoichiometry
S MATRIX 1 0 0 0 1 2 0 1 0 T MATRIX 0 0 0 1 2 1 1 0 1
Source Stoichiometry + Target Stoichiometry + V Matrix
S MATRIX 1 0 0 1 0 1 T MATRIX 0 0 1 2 1 0 V MATRIX 1 0 0 2 0 1 REVERSIBLE 0 1
This format includes all information about the reactions.
Human Readable
A --> B + C B + C <--> 2B
This format includes all information about the reactions. Note that the two reactions are represented on separate lines, despite both having the complex in common.
TO DO: Finalise modulators in file formats and describe them here.
Terminology and notation
Nonnegative vector, positive vector
An vector is termed nonnegative (and we write ) if each of its components is nonnegative, i.e., . is termed positive (and we write ) if each of its components is positive, i.e., .
Stoichiometric subspace and stoichiometry classes
The stoichiometric subspace of a CRN is the image of the stoichiometric matrix . If the stoichiometric subspace is a linear subspace of .
Stoichiometry classes, also known as a “stoichiometric compatibility classes” of a CRN with stoichiometric matrix are the sets formed by intersection of cosets of the stoichiometric subspace with the nonnegative orthant. I.e. given the stoichiometry class of is the set . Given a system of the form with and such that is forward invariant, the stoichiometry classes are clearly forward invariant sets for the system. A stoichiometry class is termed “nontrivial” if it includes a positive vector (i.e., it intersects ).
Stoichiometry classes are bounded if and only if ker includes a positive vector; equivalently if and only if im includes a nonnegative and nonzero vector. A bounded, nonempty stoichiometry class is an invariant compact polyhedron, and so must include an equilibrium by the Brouwer fixed-point theorem.
Compatible vectors/equilibria
Given a system , two vectors are “compatible” if they belong to the same stoichiometry class. In particular are compatible equilibria if , and .
Reaction system with arbitrary inflows and outflows
Also known as a “fully open system”. Consider a reaction system with . Then given any nonnegative vector and any function with derivative which is positive and diagonal on , the new system will be termed “the system with arbitrary inflows and outflows”. If a claim is made for “the system with arbitrary inflows and outflows”, this means that the claim must hold true for all nonnegative vectors and functions satisfying the assumptions above.
Siphon
A siphon of a CRN is a set of chemical species such that if the concentrations of each species of is zero, then no reaction is able to produce any species of (for general kinetics). In other words if the concentrations of species in a siphon are set to zero, then they remain zero for all time. Given an ordered set of substrates, we can regard as siphon as a nonempty, proper, subset of . Corresponding to such an is a face of defined by . It is easy to see that if is a siphon, then is invariant under weak assumptions on the reaction kinetics.
MPNE
“Multiple positive nondegenerate equilibria”: when there are two or more positive equilibria on a stoichiometry class and the reduced Jacobian determinant is nonzero at each of these equilibria.
Persistence-type conditions
Persistence condition 1 (PC1)
The boundary of includes no equilibria other than the origin. This can be claimed for general kinetics if, for example, the system has no siphons.
Persistence condition 2 (PC2)
No nontrivial stoichiometry class includes any equilibria on the boundary of . This can be claimed for general kinetics if, for example, no nontrivial stoichiometry class intersects a face of corresponding to a siphon. A CRN satisfying PC2 can be termed structurally persistent.
Persistence condition 3 (PC3)
No initial condition in has omega limit set intersecting the boundary of . (This is persistence in the usual sense.) This may occur for mass action kinetics even when some nontrivial stoichiometry class intersects a face of corresponding to a siphon. The persistence conjecture states that all weakly reversible CRNs with mass action kinetics are persistent.
Injectivity-type conditions
Injectivity condition 1 (IC1)
The system satisfies condition IC1 if: and implies . In other words, the vector field cannot take the same value at two distinct compatible vectors both in the interior of . This can be phrased briefly as “the system forbids multiple compatible positive equilibria”.
Injectivity condition 1a (IC1a)
The system satisfies condition IC1a if: and ], imply .
Injectivity condition 1'' (IC1'')
The system satisfies condition IC1'' if: , , and together imply that and in fact both and belong to the same facet of . In other words, if the vector field takes the same value at two distinct compatible vectors, then they must both be on the boundary of and in fact on the same facet of .
In particular, the practically important conclusions are: (i) any positive equilibrium is the unique equilibrium on its stoichiometry class; (ii) if a stoichiometry class includes a boundary equilibrium , then all equilibria on the stoichiometry class of must belong to a single facet of .
Injectivity condition 2 (IC2)
The system satisfies condition IC2 if the associated system with arbitrary inflows and outflows is injective on . In particular it has at most one equilibrium in . A corollary is that the original system cannot have MPNE.
Injectivity condition 2'' (IC2'')
The system satisfies condition IC2'' if the associated system with arbitrary inflows and outflows is injective on . In particular it has at most one equilibrium in .
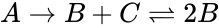
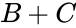 in common.
in common.
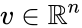 is termed nonnegative (and we write
is termed nonnegative (and we write 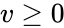 ) if each of its components is nonnegative, i.e.,
) if each of its components is nonnegative, i.e., 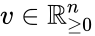 .
. 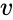 is termed positive (and we write
is termed positive (and we write 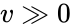 ) if each of its components is positive, i.e.,
) if each of its components is positive, i.e., 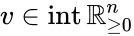 .
.
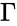 . If
. If 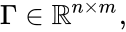 the stoichiometric subspace is a linear subspace of
the stoichiometric subspace is a linear subspace of 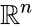 .
.
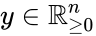 is the set
is the set 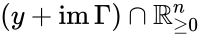 . Given a system of the form
. Given a system of the form 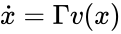 with
with  and such that
and such that 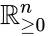 is forward invariant, the stoichiometry classes are clearly forward invariant sets for the system. A stoichiometry class is termed “nontrivial” if it includes a positive vector (i.e., it intersects
is forward invariant, the stoichiometry classes are clearly forward invariant sets for the system. A stoichiometry class is termed “nontrivial” if it includes a positive vector (i.e., it intersects 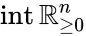 ).
).
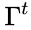 includes a positive vector; equivalently if and only if im
includes a positive vector; equivalently if and only if im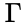 includes a nonnegative and nonzero vector. A bounded, nonempty stoichiometry class is an invariant compact polyhedron, and so must include an equilibrium by the Brouwer fixed-point theorem.
includes a nonnegative and nonzero vector. A bounded, nonempty stoichiometry class is an invariant compact polyhedron, and so must include an equilibrium by the Brouwer fixed-point theorem.
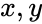 are compatible equilibria if
are compatible equilibria if 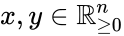 ,
, 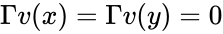 and
and 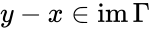 .
.
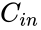 and any
and any 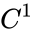 function
function 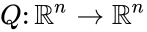 with derivative
with derivative 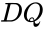 which is positive and diagonal on
which is positive and diagonal on 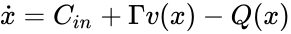 will be termed “the system with arbitrary inflows and outflows”. If a claim is made for “the system with arbitrary inflows and outflows”, this means that the claim must hold true for all nonnegative vectors
will be termed “the system with arbitrary inflows and outflows”. If a claim is made for “the system with arbitrary inflows and outflows”, this means that the claim must hold true for all nonnegative vectors  satisfying the assumptions above.
satisfying the assumptions above.
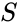 such that if the concentrations of each species of
such that if the concentrations of each species of 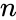 substrates, we can regard as siphon
substrates, we can regard as siphon  . Corresponding to such an
. Corresponding to such an 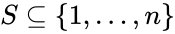 is a face
is a face 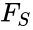 of
of 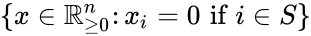 . It is easy to see that if
. It is easy to see that if 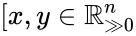 and
and ![{\displaystyle y-x\in \mathrm {im} \,\Gamma \backslash \{0\}]}](https://wikimedia.org/api/rest_v1/media/math/render/png/39ecfe9b5d5cb63ea89273ac595c7616a2cb6497) implies
implies 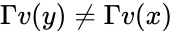 . In other words, the vector field cannot take the same value at two distinct compatible vectors both in the interior of
. In other words, the vector field cannot take the same value at two distinct compatible vectors both in the interior of 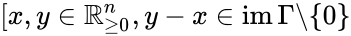 and
and 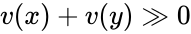 ], imply
], imply 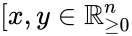 ,
, 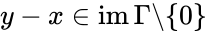 , and
, and ![{\displaystyle \Gamma v(y)=\Gamma v(x)]}](https://wikimedia.org/api/rest_v1/media/math/render/png/7b3ced12e6445d2a71f9cf0229fbf39bddbea570) together imply that
together imply that 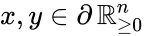 and in fact both
and in fact both 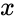 and
and 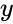 belong to the same facet of
belong to the same facet of 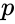 , then all equilibria on the stoichiometry class of
, then all equilibria on the stoichiometry class of 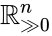 . In particular it has at most one equilibrium in
. In particular it has at most one equilibrium in